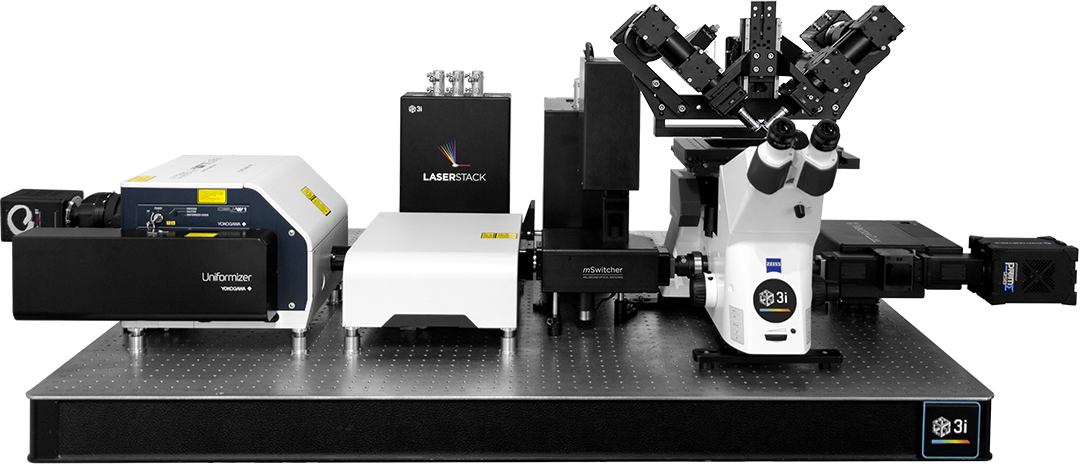




Environmental Control
Stage top and cage incubators
Temperature, gas and humidity
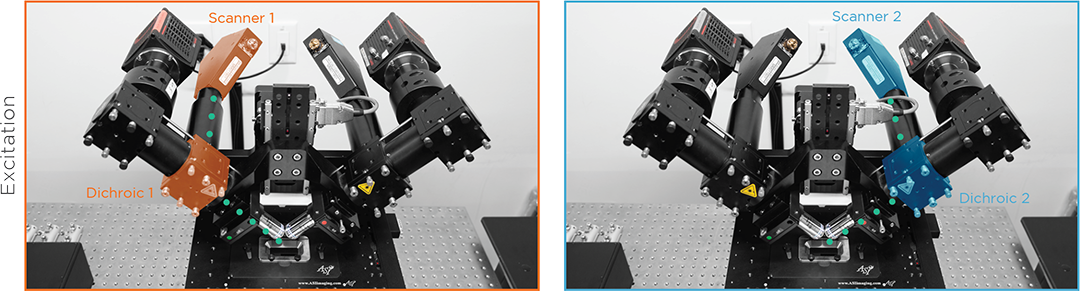
Light Sheet Scanners
Objective Piezos

Motorized SPIM Z Drive

High-speed MEMs mirror or cylindrical lens scanners
95% QE low read noise
Large field-of-view options
150µm or 300µm piezos for long-term high-speed 3D sampling
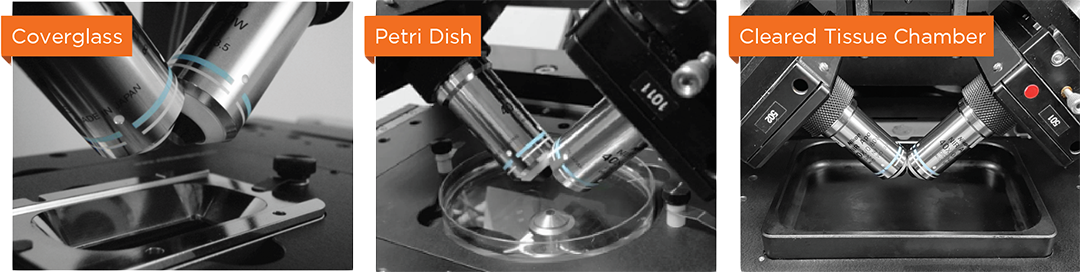
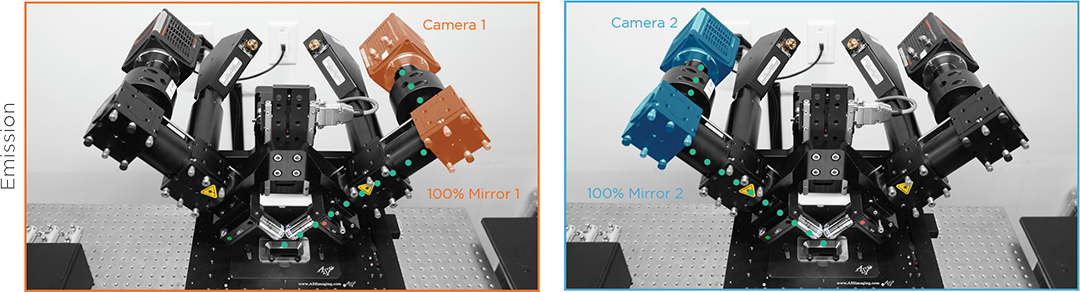
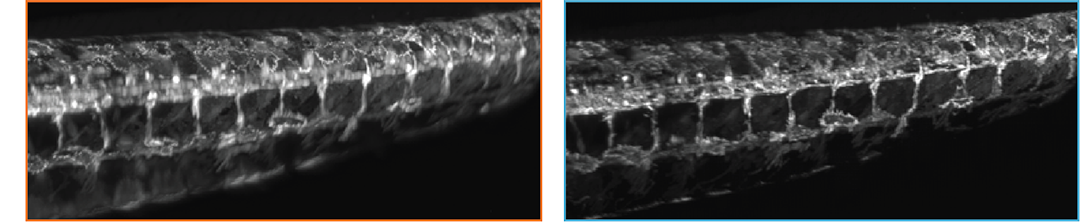
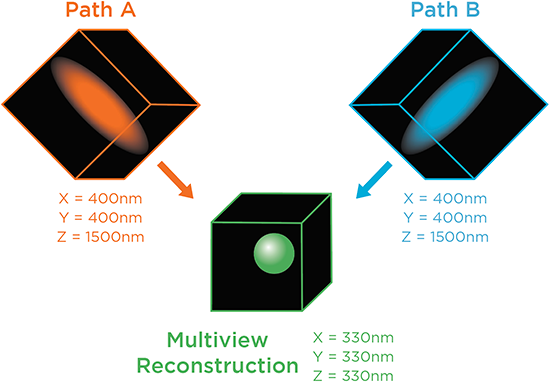
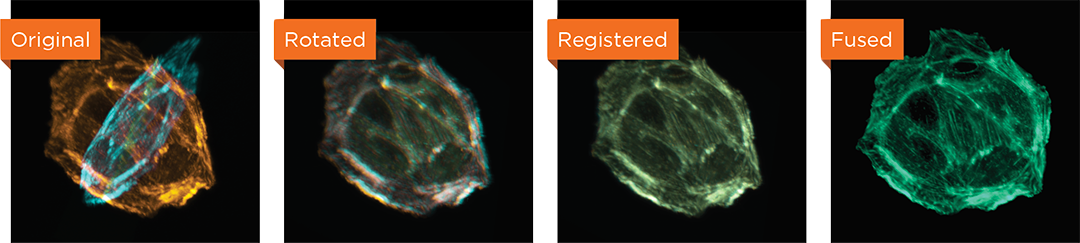
Water and multi-immersion dual-view optimized objective pairs
Motorized objective, condenser and path selection
Autofocus (Definite Focus 3)
PSF-optimized objectives
High precision multi-position and vertical montage capture
Scan-Optimized X,Y Stage



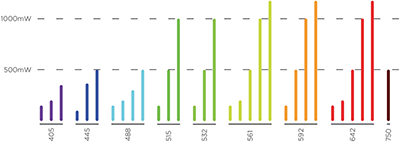


LaserStack
Fiber Switcher
Additional Light Path

Millisecond timing and trigger
Control of multiple devices
Modular laser combiner
Up to eight lasers
Up to four fiber outputs
Millisecond path switching
Add cameras, photomanipulation devices,
spinning disk confocal and more
High-precision movement of sample during long-term experiments

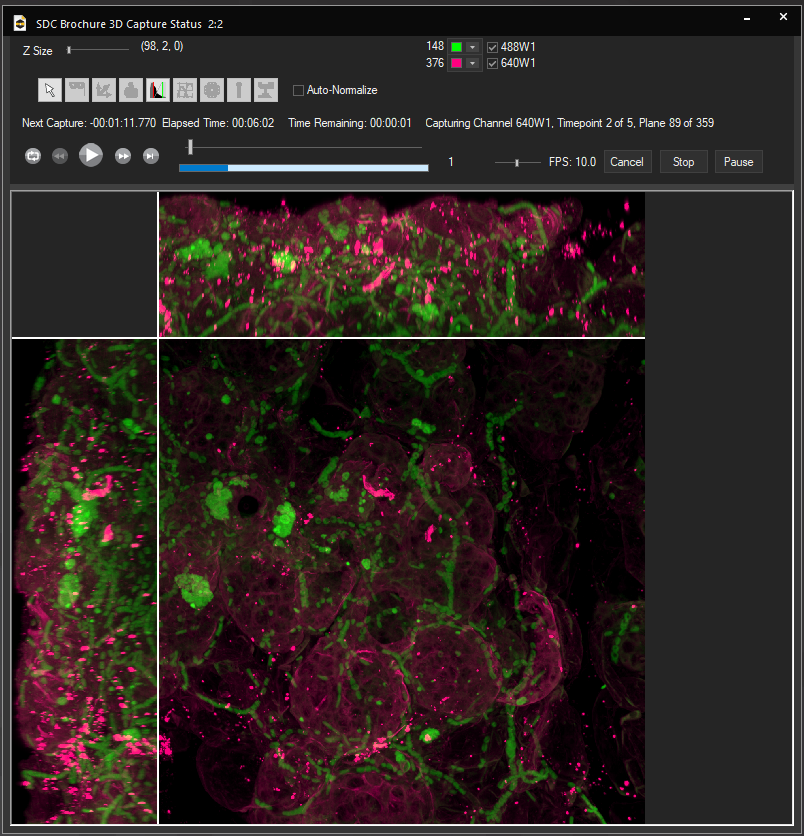
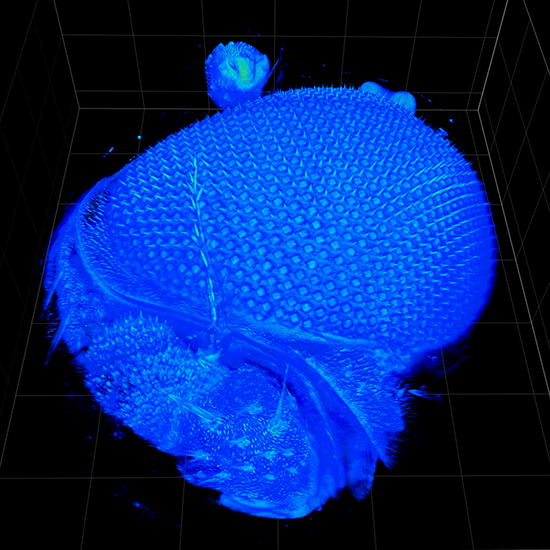
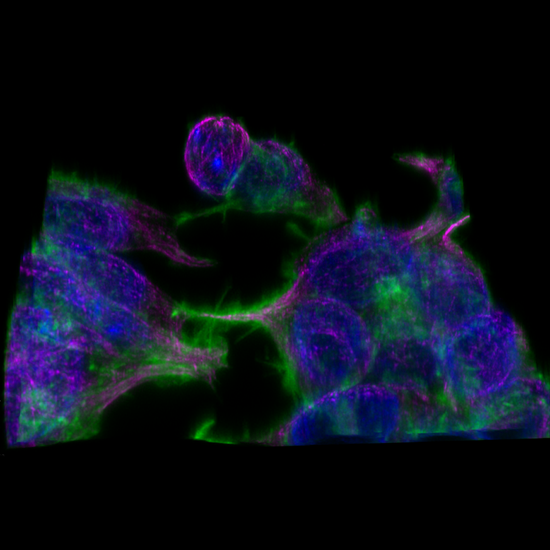
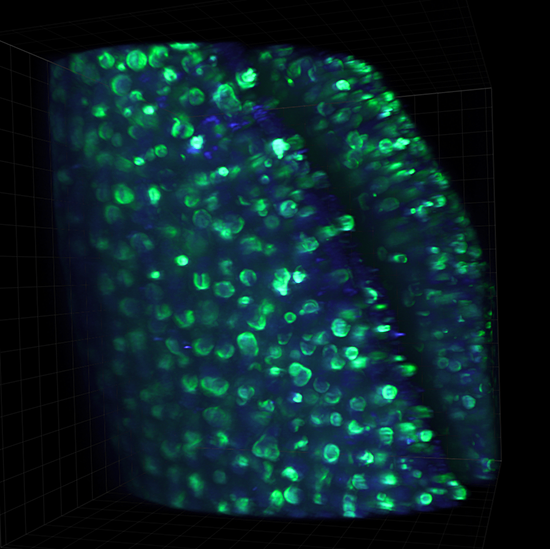
Live cell 3D confocal imaging
Super-resolution dual microlens disk

Ablate
Laser ablation system
355nm and 532nm
sCMOS Cameras


Vector2
Vector3
Objectives
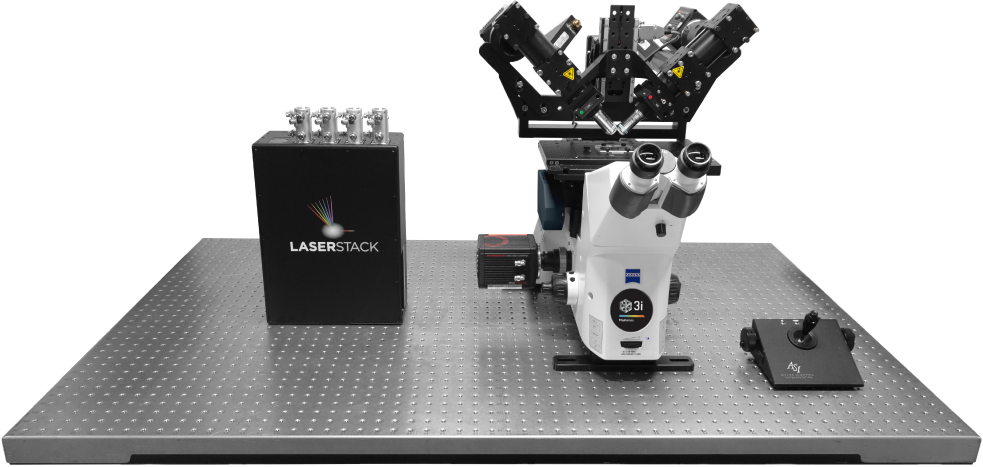
Fully Automated
Research Microscope
Epi mounted or camera port
mounted photomanipulation
Modular high-speed X,Y scanner
Epi-mounted TIRF & photomanipulation
Spinning TIRF with expansive FN22 FOV
Liquid light guide input for LED light source