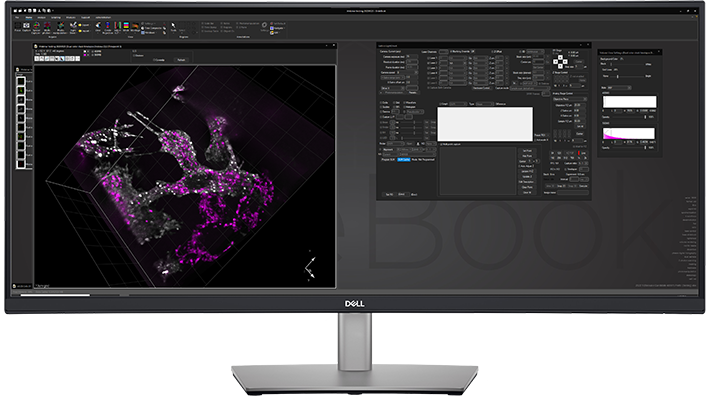

Beam Steering
Fast X and Z galvanometers for rapid volumetric captures
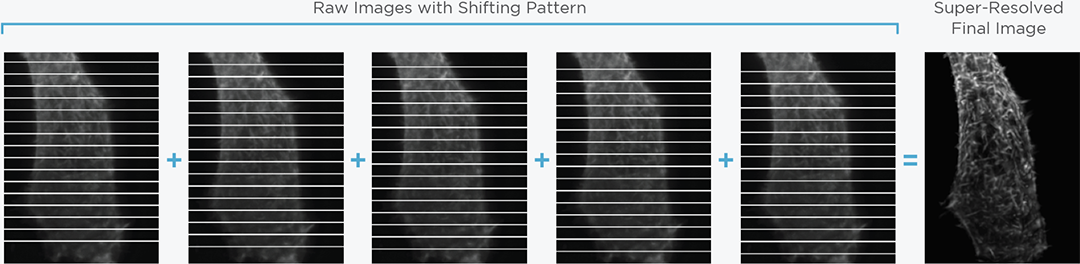
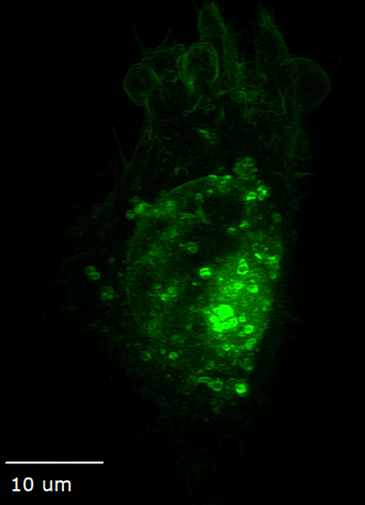
Structured illumination for super-resolution imaging
Sensitive Cameras

Environmental Control

95% QE low read noise sCMOS detectors
Fast, sensitive spectral separation
Up to 2 cameras
Sample chamber and objective heating
Digital temperature, gas, and humidity control
Motorized Light Sheet Selection


Upright Microscope Configuration
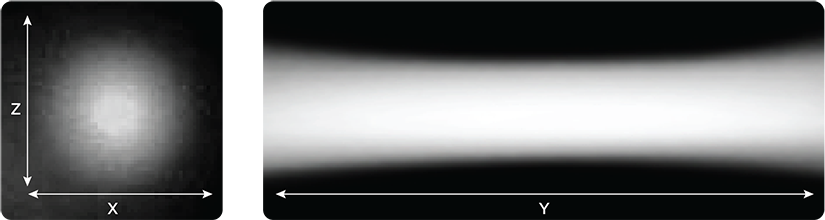
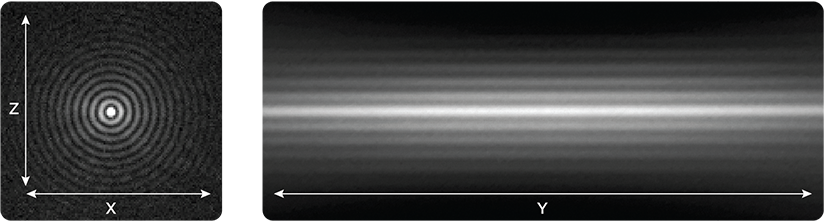
Digital Light Sheet Formation

Standard glass sample mounting for cells and tissue
Custom sample mounting on other materials
Highest resolution, water-dipping objective pair
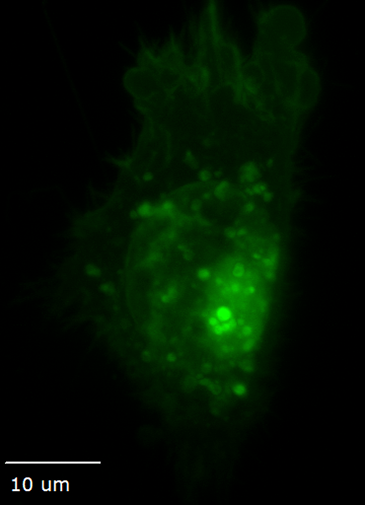
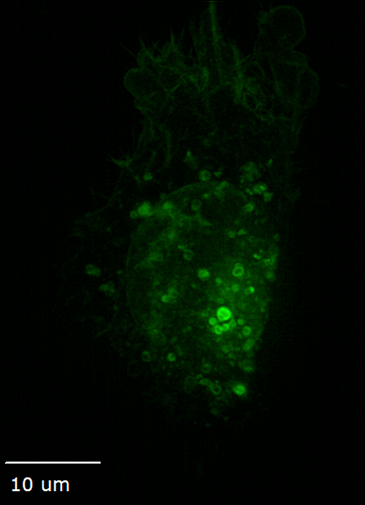
Ideal for live cells, organoids, tissue, and embryos
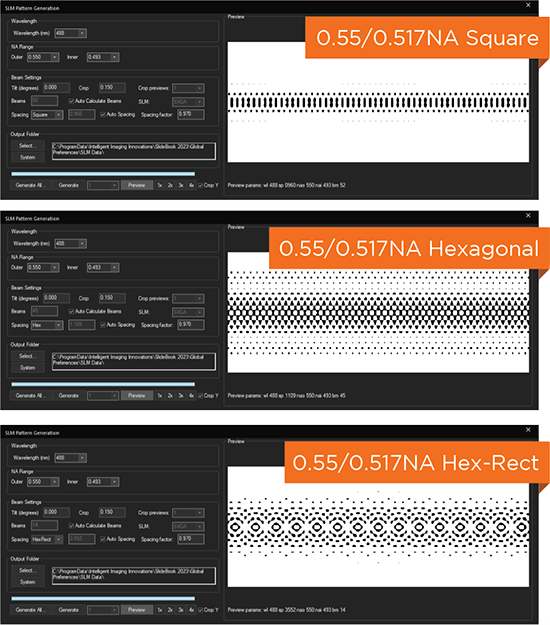
Beam structuring via spatial light modulator (SLM)
Square, Hexagonal, Bessel & other patterns
Adjustable light sheet length and thickness for a variety of samples, without realignment

Widefield Imaging Path
Inverted widefield path for easy sample finding
Brightfield LED & epi-fluorescence LED options
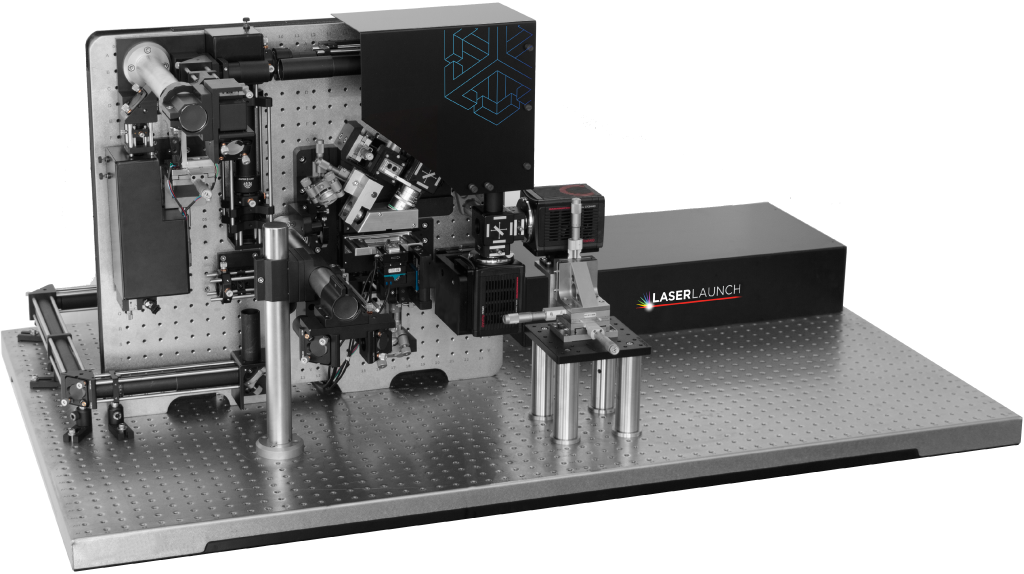
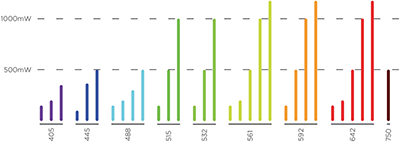
LaserLaunch Modular Laser Combiner