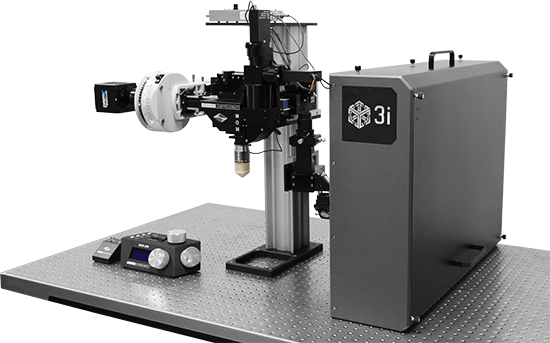
2P Holography | Maximum Power
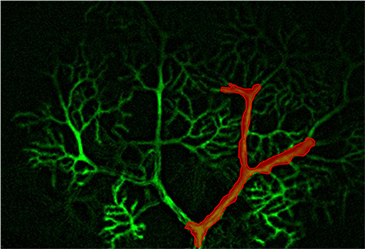
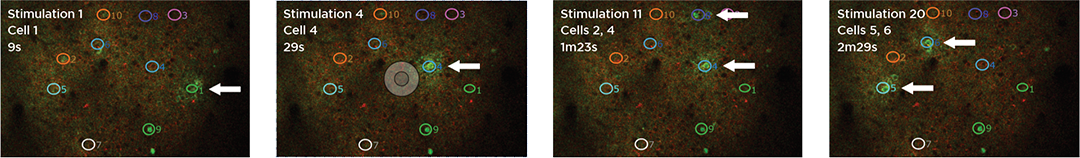
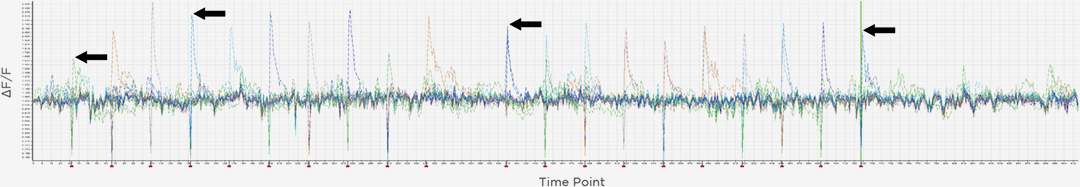
3D Holographic Stimulation
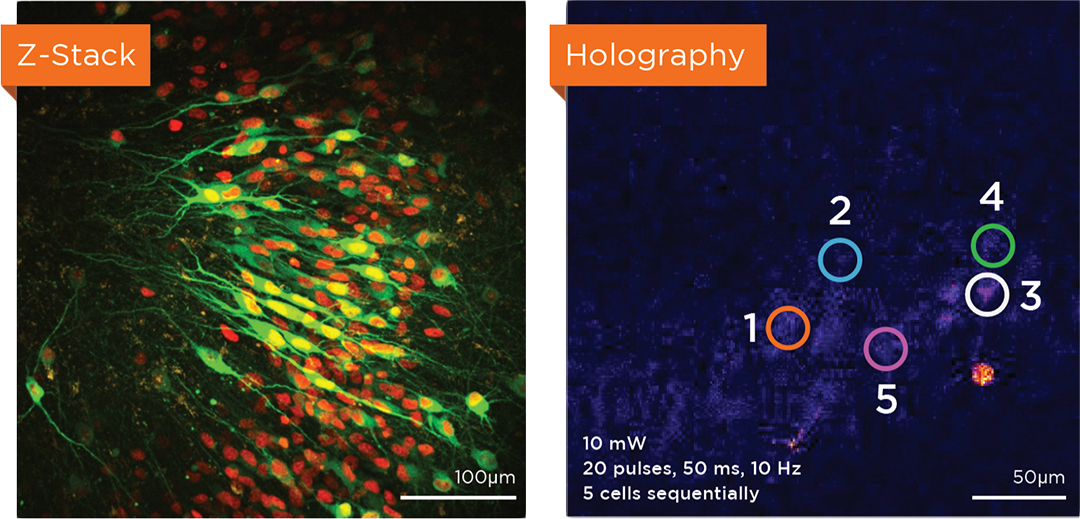
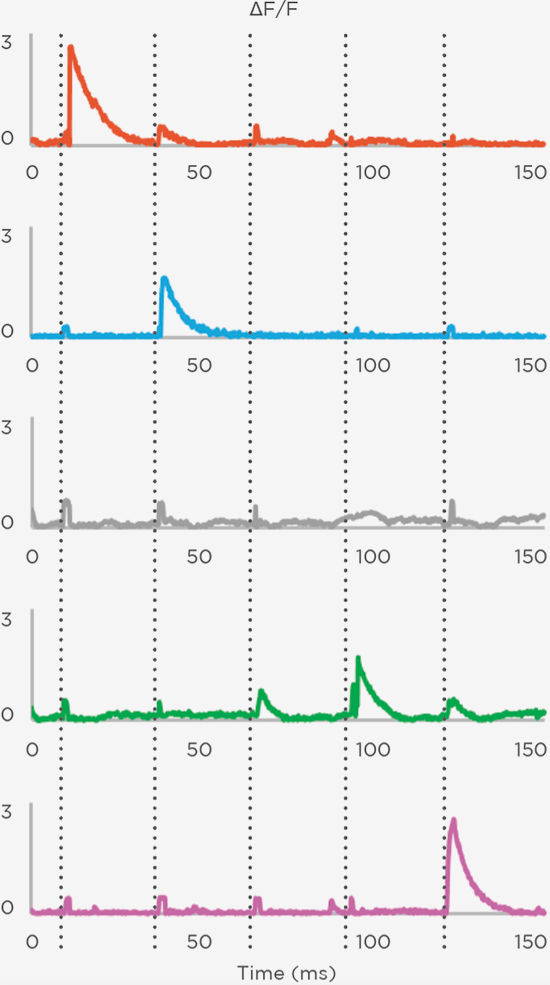
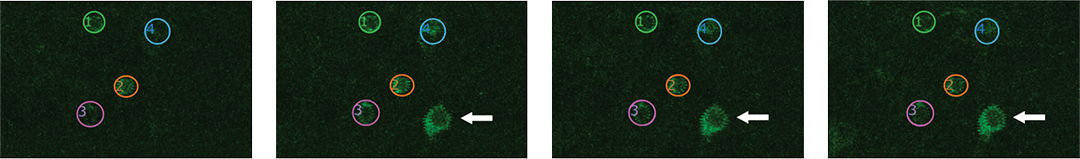
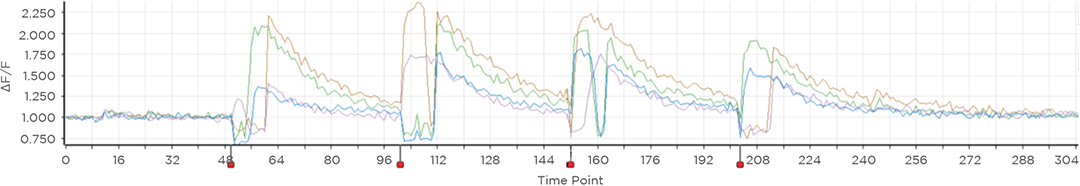
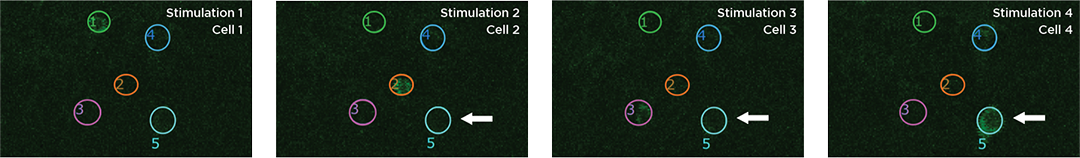
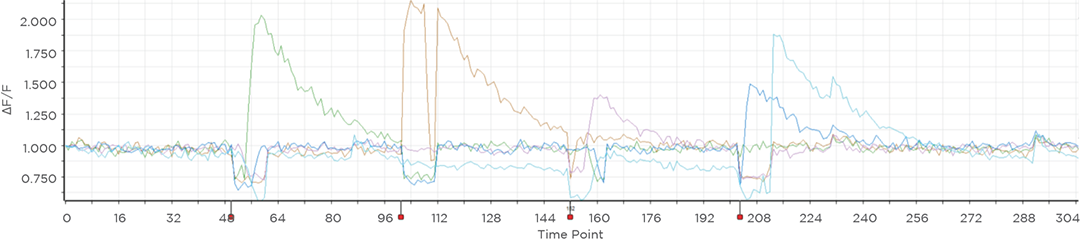
Proprietary software based on iterative Fourier transform algorithms (IFTA) simultaneously generate multiple hand-drawn regions of interest at any position in sample space.
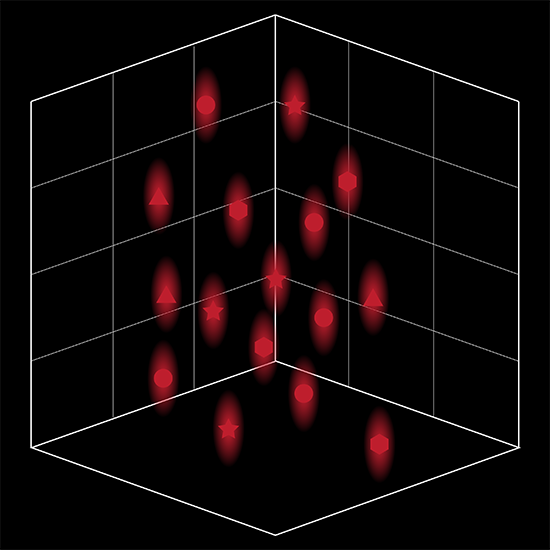
SlideBook’s exclusively licensed libraries generate holographic patterns at multiple Z positions with corrections for power discrepancies inherent to the use of SLMs and each pattern can be assigned a specific power density.