for acquisition and analysis
SlideBook digital microscopy software advances research microscopy through the entire experimental process. By managing everything from instrument control to image processing and data analysis, SlideBook allows scientists to focus on investigation rather than instrumentation. SlideBook comes standard with drivers to control hundreds of instruments in and around the microscope. Online, data is acquired in a native-3D format over time, color and specimen locations in customizable experiment protocols. Offline, data can be analyzed by a wide variety of tools for image processing including mathematical operations, statistics functions, analysis scripting and import/export to/from other microscopy software. Additional modules are available for special applications ranging from deconvolution to photomanipulation, holography, light sheet, adaptive optics and more.
SlideBook Features
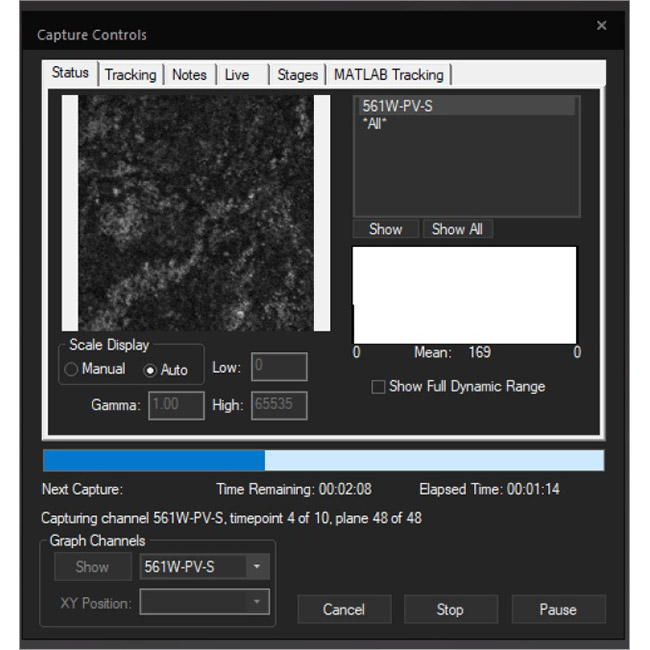
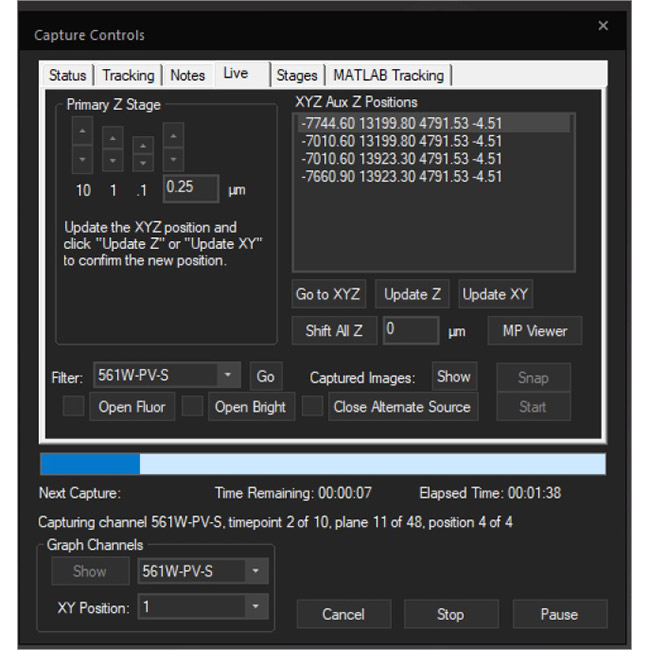
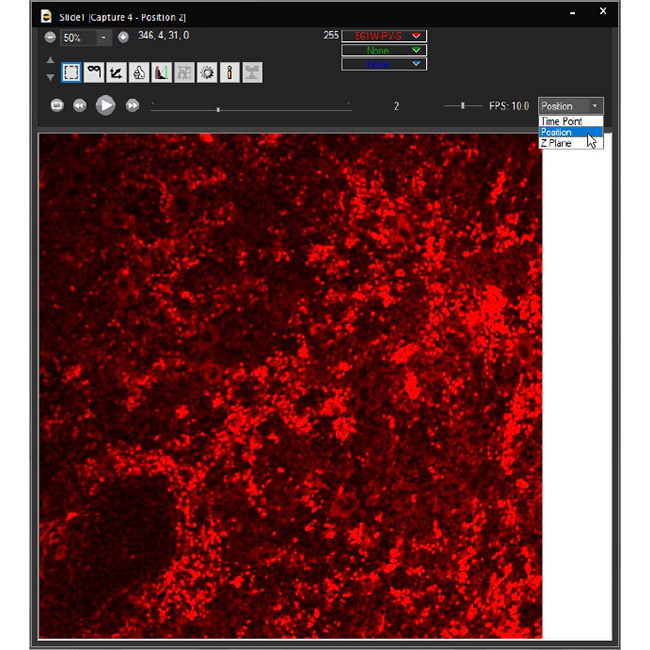
Examine datasets during and after acquisition with SlideBook’s 3D Capture Status and Multipoint Viewer. 3D Capture Status supports volumetric projection (via max-intensity projections) to observe dynamics during an experiment by examining more than one plane at a time. The Multipoint viewer allows for examination of multiple XYZT positions during acquisition, and post-acquisition the user may review all the XYZT positions in a capture.
3D Capture Status
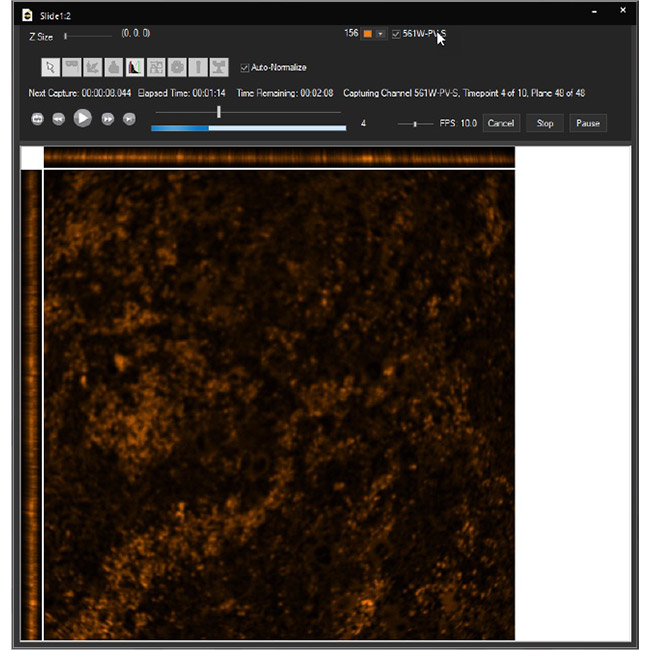
Multipoint Viewer
The .sldy file format is an open-source-friendly file format designed for very large data sets (from 100GB to >1TB). The .sldy file is a distributed file format that points to a directory where each captured 2D or 3D image is stored in an independent file (by channel and timepoint) alongside metadata files describing the capture and microscope parameters. The distributed nature makes image manipulation operations – such as removing a channel or a selection of time points – nearly instantaneous. Images, masks, and histograms are stored as Python-compatible NumPy arrays, and the metadata is stored in the open-source YAML format, enabling users with coding experience to process images directly in Python. The default .sld file format is still available and appropriate for many users.
.sld is a single file that contains all images and metadata
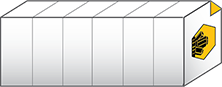
.sldy is an open-source-friendly collection of individual image and metadata files in a directory. The .sldy file format allows for much faster execution of certain file operations, such as deleting a single image or timepoint.
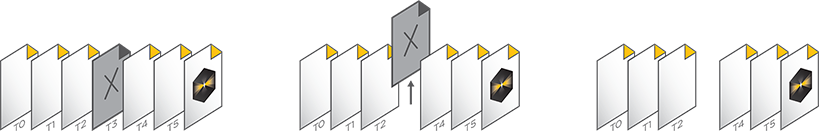
The .sldy file format is also natively compatible with Python, allowing direct access to image data without the need for a special reader.

SlideBook supports compression of the .sldy file format (called .sldyz) with the Zstandard compression algorithm. Zstandard is a lossless algorithm that compresses very large files with a balance of speed and compression ratios that outperforms standard lossless algorithms. Each NumPy image file is compressed individually, so the structure remains the same, but the overall size of the directory is smaller. Compressed files can be opened natively in SlideBook with no need to decompress them.
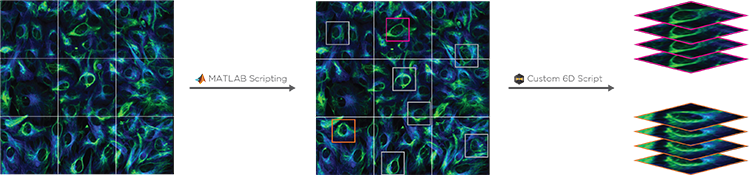
Conditional and Hierarchical Capture allows a MATLAB script to control experimental workflows. For example, a user can automate the selection and imaging of certain cells at higher magnification after a low-mag pass, or to stop a timelapse image after a particular event has been observed. By coupling with 6D Multicapture, users can customize highly specific and complex automated experiments.
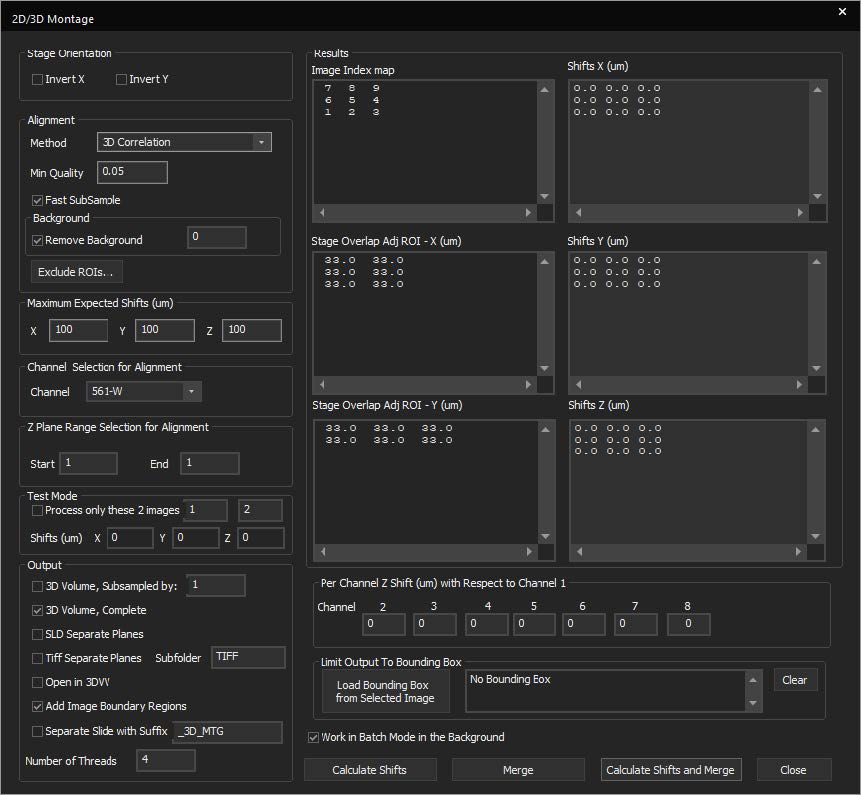
SlideBook features a montaging tool for stitching 2D, 3D, and 4D data. This allows for better handling of large datasets in a constrained memory environment. New features include:
- Test alignments on a subsection of the overall image
- Limit the alignment to a smaller Z-range in 3D and 4D datasets
- Subsample before stitching
- Output the montaged data to a new file
- Limit the number of CPU threads and run the job in the background to free resources for other operations
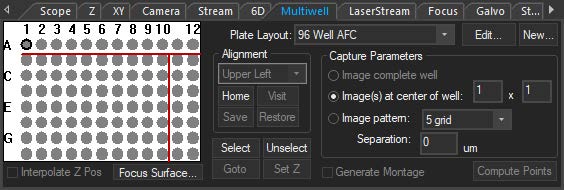
The multiwell interface is incorporated into the focus window to allow for easier plate alignment and selection of wells for imaging. The selected wells are seamlessly integrated into the normal SlideBook workflow, thus reducing the amount of time, mouse-clicks, and learning curve to perform multiwell imaging. There are several pre-set well plate layouts, and users can also define custom well plates of varying sizes up to 384 wells.
SlideBook’s interface for multiphoton systems assembles all frequently-used controls and status displays into a single window. The console features an advanced, intuitive tool for adjusting laser power delivery for different depths incorporating dynamic signal feedback.
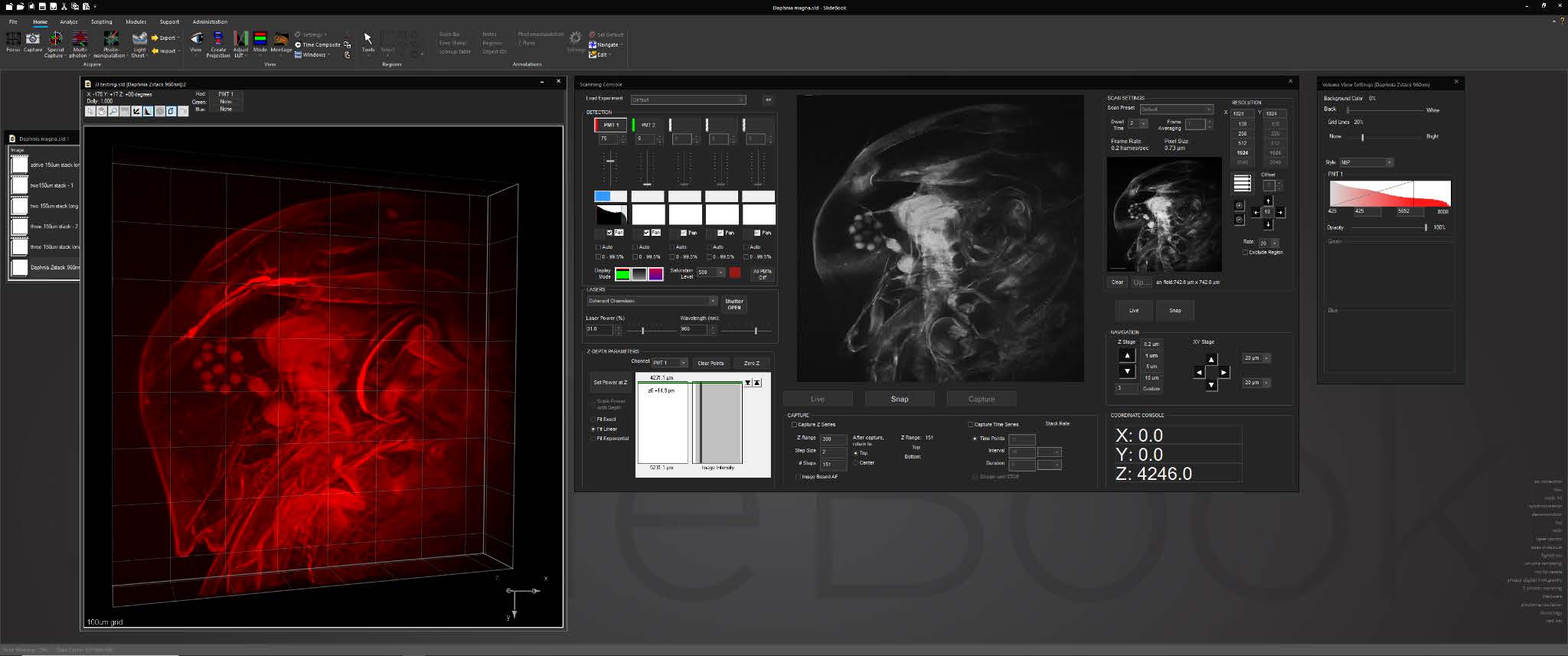
For cleared tissue light sheet imaging, a console for AxL Cleared Tissue LightSheet is intuitive and feature rich, with features such as 2D and 3D prescan, ROI selection, automated lightsheet pattern generation, and axial chromatic aberration correction.
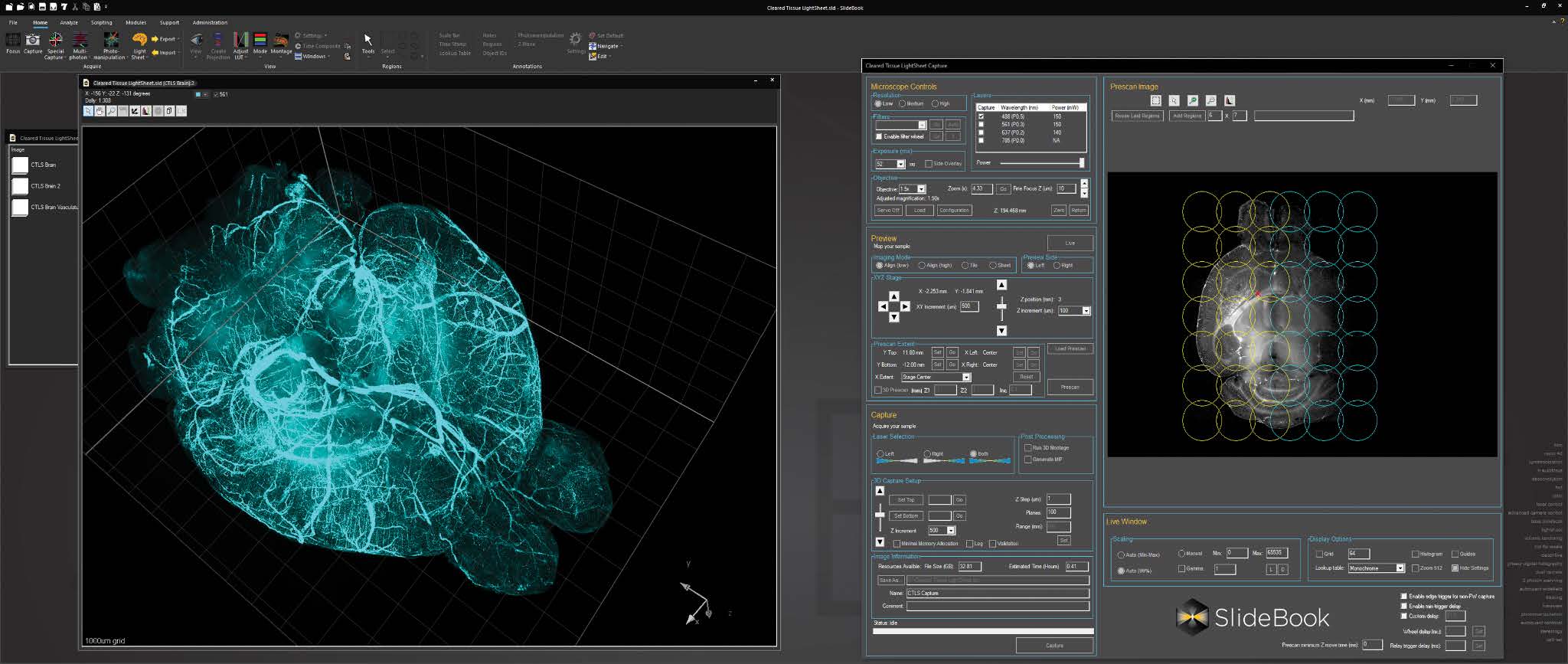
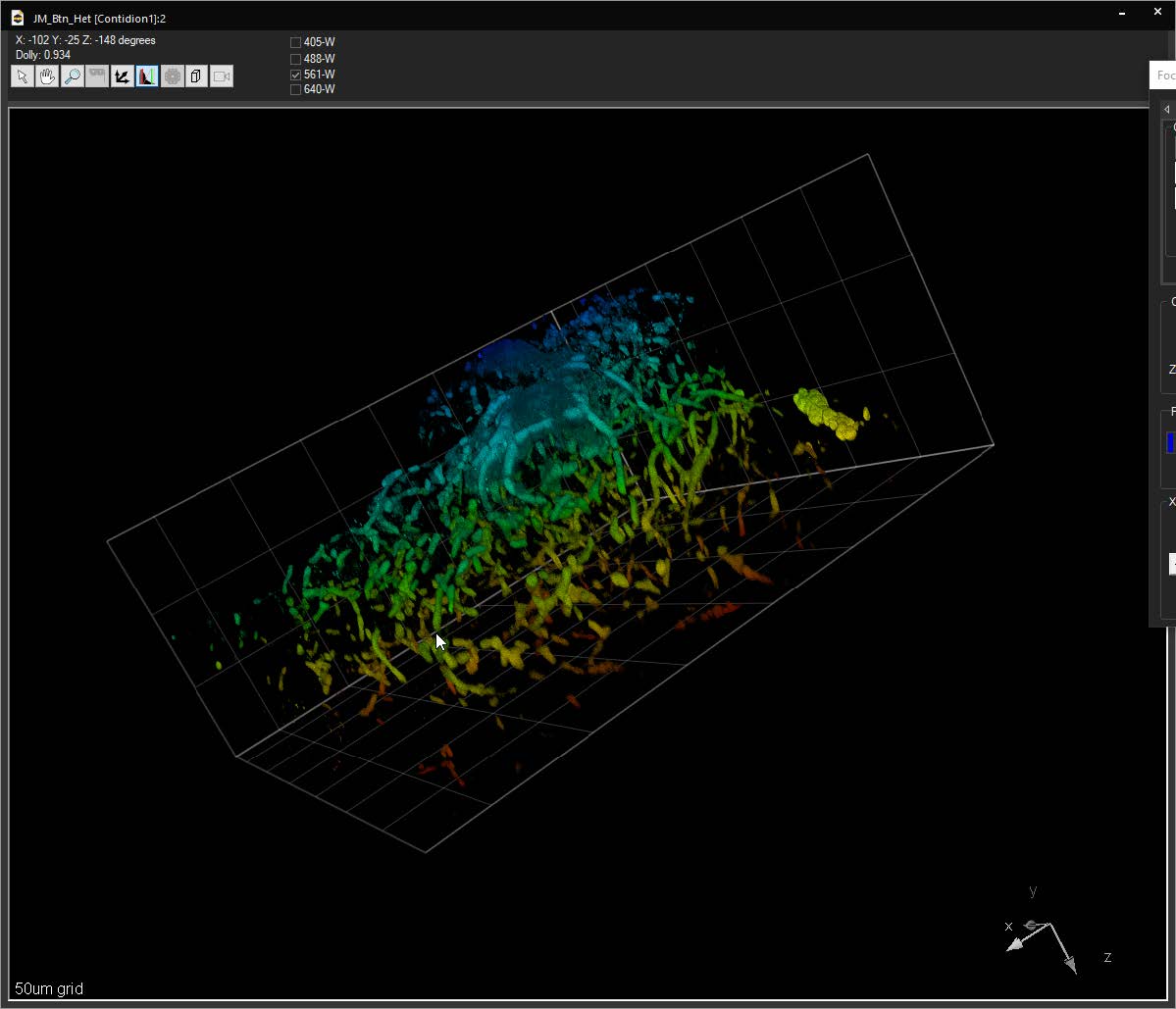
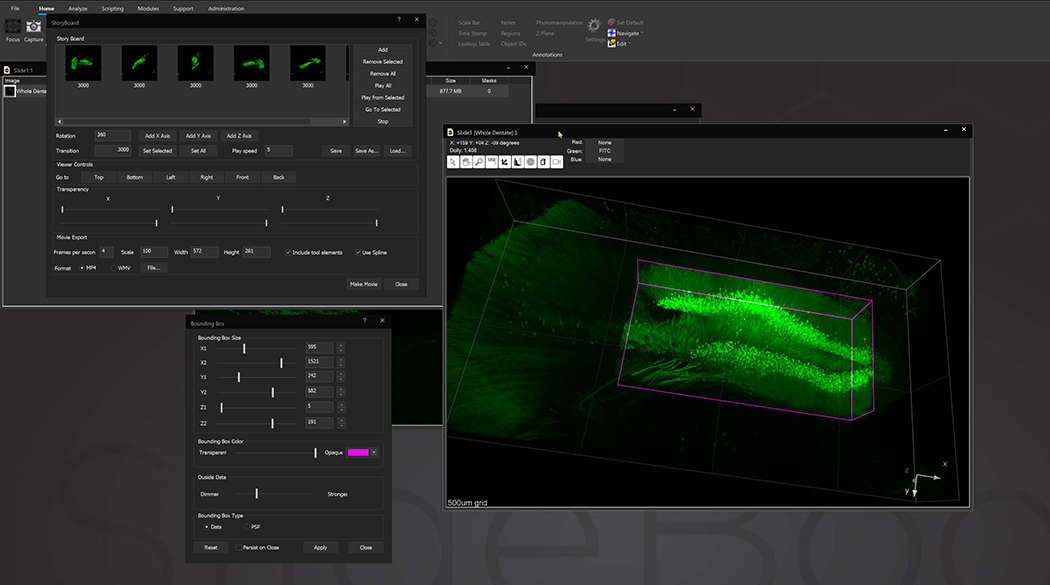
Bounding box in the 3D Volume View allows for cut-away viewing of projected volumes in all three dimensions and selection of subsets of the data for other operations. Users are also able to change the transparency and opacity of portions outside the bounding box. The bounding box functionality can be extended for use in making movies, calculating point spread functions and montaging.
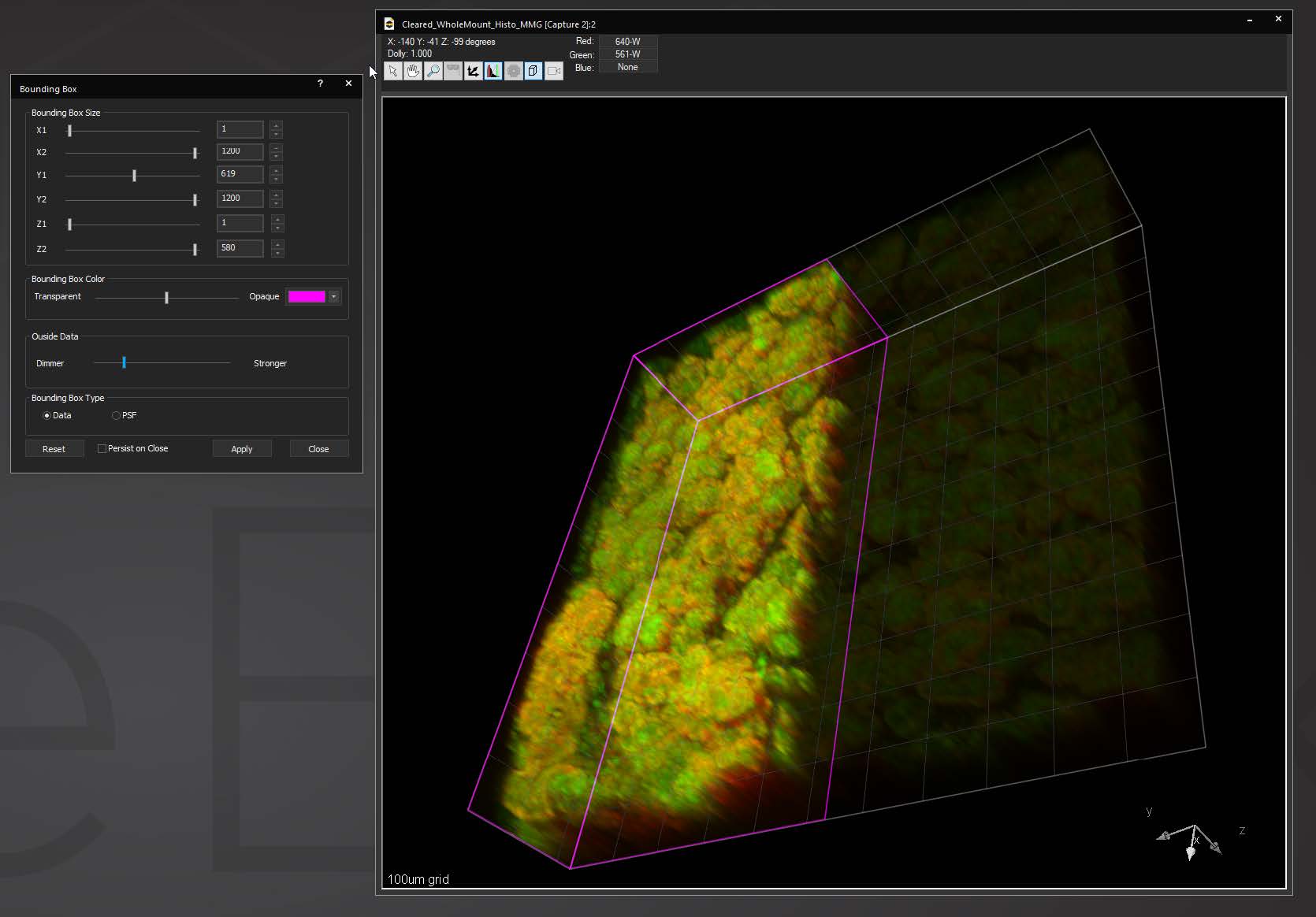
StoryBoard is an all-new way to make movies from 3D and 4D datasets utilizing keyframes and custom framerates. Movie templates can be saved and re-used for multiple datasets. Incorporating spline interpolation, StoryBoard quickly renders beautiful and smoothly animated movies from large datasets.
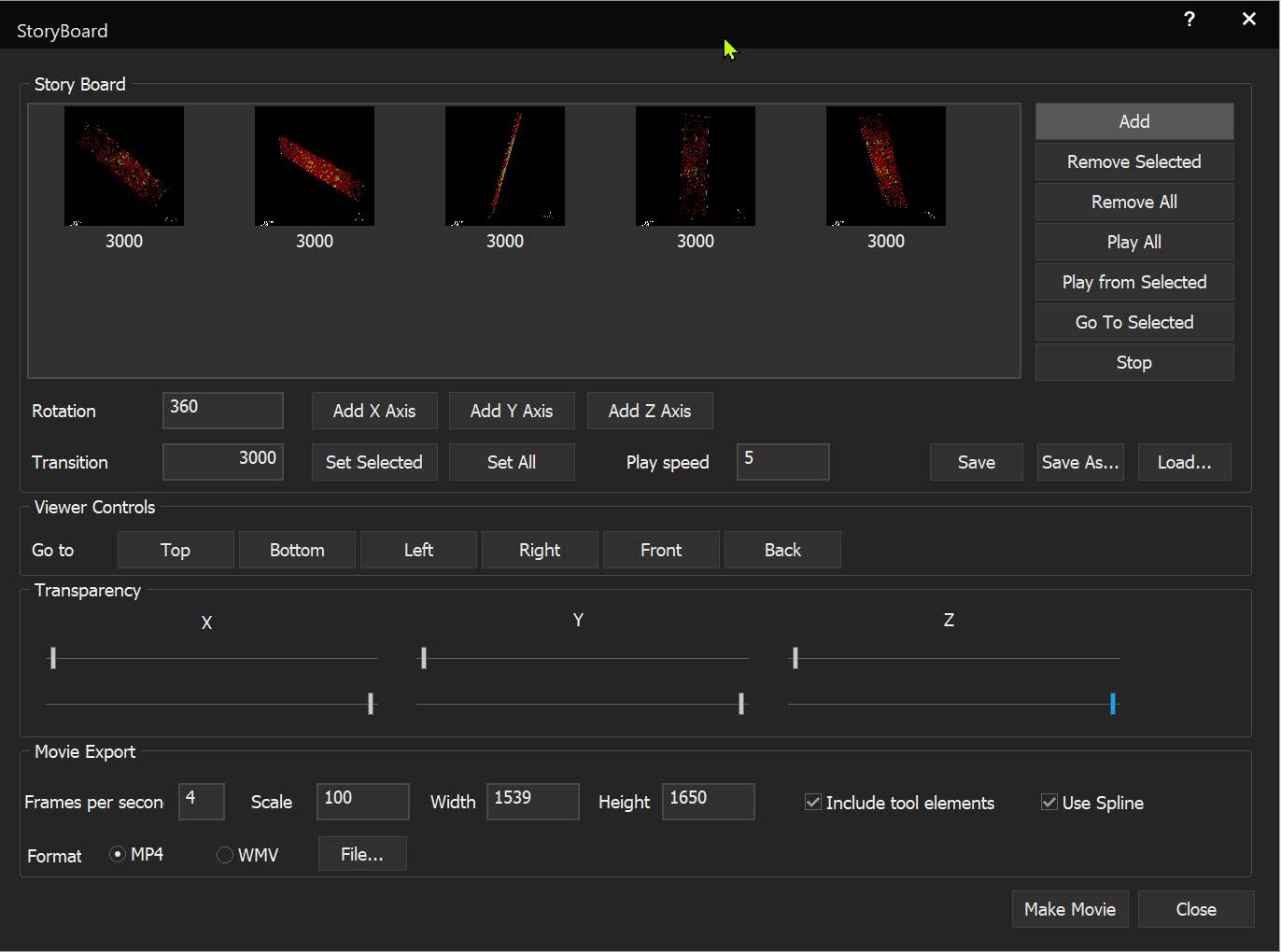
SlideBook can link to ImageJ lookup tables (LUTs) and display 3D volumes with different colors depending on depth. The same lookup tables can be applied to other captures using the Pseudocolor Mode on a per-filter basis.
User-selectable color schemes with dozens of different options including light, dark and black modes.
Focus Surface allows users to correct for non-planar deformities by selecting multiple points of focus and interpolating a surface across them to automatically adjust focus as the system moves between points, wells, or tiles in a montage. This is helpful in situations where tissue may be warped or undulating from fixing and dehydrating, or where a multiwell plate is deformed due to slight imperfections in the plastic.

NVIDIA CUDA leverages parallel processing on the GPU to greatly speed up operations by executing many commands at the same time. Operations that have been CUDA-optimized in SlideBook 2022 include:
- Deconvolution (all modalities: No Neighbors, Nearest Neighbors, Constrained Iterative, Big Data, MLS Decon)
- MLS Localized Cross-correlation
- MLS POI Registration
- Adaptive Optics
- Software-based autofocus
- 3D montage
- Image deskew and rotations
- Max-intensity projections
Capabilities

Capture
SlideBook is able to drive hundreds of devices including microscopes, stages, lasers, wheels, piezos, scanners, shutters and many more via serial commands and TTL pulses. SlideBook is capable of robust data acquisition including z stacks in a native 3D format, time lapse, multiple color channels, multiple stage locations and fluorescence lifetimes. Configuration settings of acquisition channels allow capture of data easily and logically without requiring setting of each device for each experiment.
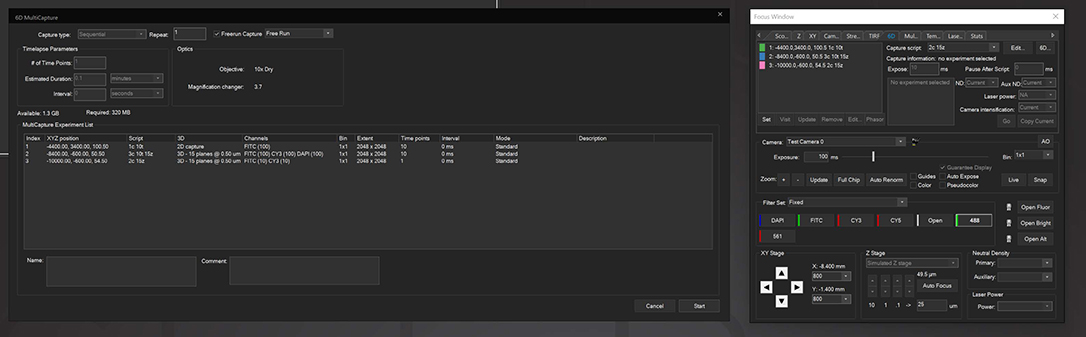
6D Multicapture
SlideBook allows users to save capture parameters as defined “Capture Scripts” that can be individually applied to different XYZ positions, allowing for vastly increased flexibility in imaging different locations under different conditions. 6D Multicapture can also be set to “Freerun” mode for high-speed multichannel and multidimensional streaming.

View
See and inspect specimen data through any number of portals from single images to z-stacks, time lapse, color channels and 4D views. Adjust raw data with intelligent processing via flatfield correction and filtering. Deconvolve 3D data with any of six algorithms according to experimental conditions.

Scripting
Many processing commands have a macro command counterpart to automate sequences of operations and ensure that all images in each slide are processed identically. The scripting language is a simple sequence of text commands that let SlideBook know what operation to do, on which image or images to do it, and what the parameters are.
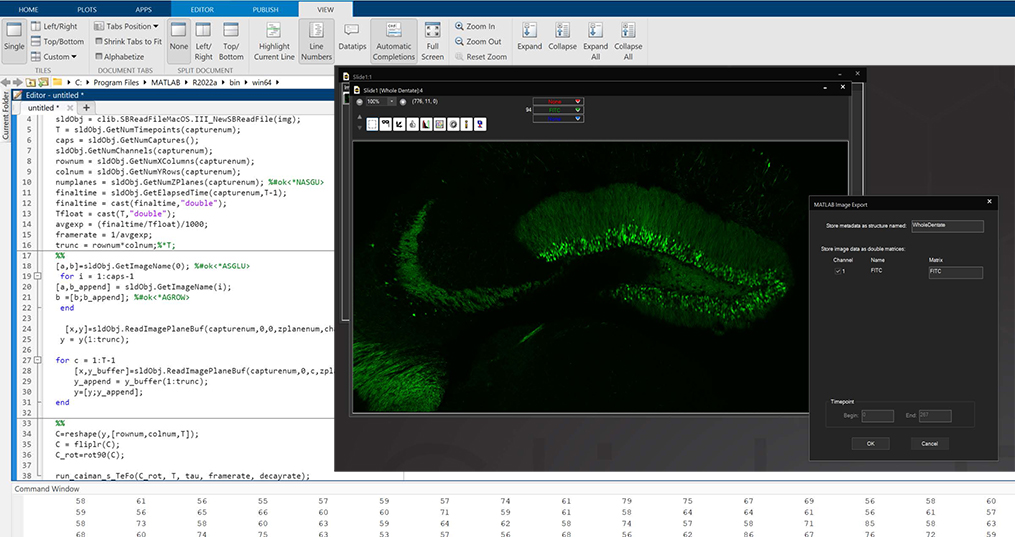
MATLAB and Python Compatibility
SlideBook is capable of directly exporting channel data and masks to MATLAB, and importing them after analysis or manipulation is complete. In addition, the .sldy file format may be accessed directly with Python, enabling expanded analysis and visualization options.
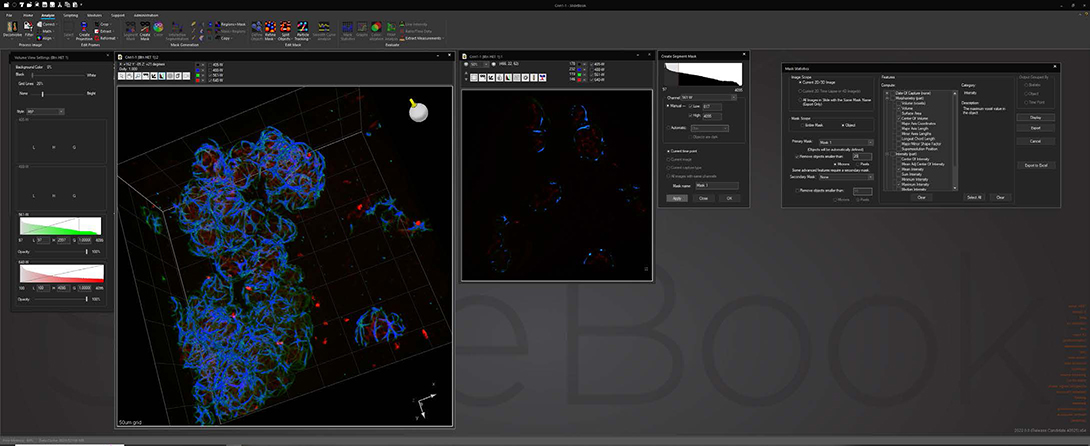
Analyze
Analyze images and extract statistical data via a wide variety of algorithms while maintaining original data integrity. Segment image data via intensity, gradient and watershed algorithms to identify areas of interest for statistical analysis. Track objects in 2D and 3D via automatic or manual delineation for analysis based on time or pathway.

Communicate
Present and export data easily as 16-bit TIFFs, 3D and 4D movies, graphs or spreadsheets. Data is directly portable to 3rd-party microscopy software and adheres to Open Microscopy Environment standards.
Modules

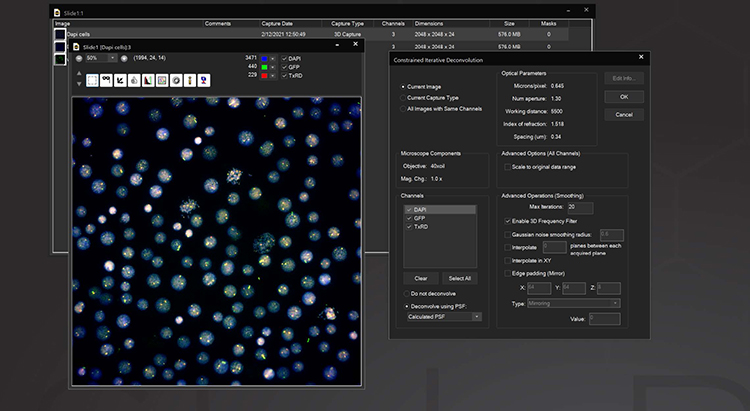
3D Deconvolution
The Deconvolution module adds both a nearest neighbor and a constrained iterative deconvolution algorithm to SlideBook. Included is an interactive guide for measuring point spread functions and a database for storing multiple PSFs. Either measured or computed PSFs can be used, with the correct PSF automatically applied to CI deconvolution without user assistance. Nearest Neighbors deconvolution is a rapid way to deblur fluorescence data using images from the plane above and below the plane of interest to correct for out-of-focus information. Constrained Iterative deconvolution is a quantitative image restoration tool. Based on the algorithm developed by David Agard at UCSF, our CI deconvolution can quantitatively reassign out-of-focus information in 3D data while improving both axial and lateral data resolution. For imaging with very low signal to noise or where measuring a PSF is not practical, the AutoQuant 3D Blind Deconvolution module may be used.

AutoQuant 3D Blind Deconvolution
This module allows the AutoQuant adaptive blind deconvolution algorithm to be run seamlessly within the SlideBook user interface. Blind deconvolution is extremely useful in situations where an objective’s point spread function cannot be accurately measured or practically captured. The PSF is iteratively reconstructed and applied to iterative reconstruction of the image data. At the expense of time, this method allows for statistically accurate data even at low signal to noise ratios.
Stereology
The Stereology module employs unbiased stereological techniques for accurate estimation of total number, volume, surface area, and length of objects in a biological structure. Stereology systematically samples 3D volumes from a series of tissue sections in a random fashion. The module integrates support for a number of stereological tools within the standard image capture framework. Montaging can be used to outline the boundaries of a structure in a tissue section and then counts performed at high magnification using standard capture tools. Support for both transmitted light and fluorescence microscopy is included, and an offline mode the permits acquisition and counting to be performed separately.

FRET
The Fluorescence Lifetime Imaging module enables measurement of fluorescence lifetimes via frequency modulation of illumination and detection signals. FLIM is a powerful tool for molecular research in living cells and can be used to measure protein proximity (FRET), polymerization, relative concentration of different molecules, separation of different labels with spectral overlap, ion concentration, and remove autofluorescence. Lifetimes are measured at the sub-nanosecond level by using unique electronic timing circuitry for synchronized phase-shifted illumination. Near single molecule detection is possible. The 3i FLIM system can be used in combination with other multidimensional imaging techniques and images orders of magnitude faster than time domain systems.
Ratio Imaging
SlideBook’s Ratio Imaging module allows calibration, acquisition, graphing, and analysis of data from ratiometric indicators such as Fura-2, BCECF, and genetically expressed Cameleons. Ratio data can be uncalibrated, calibrated using buffer solutions, or calibrated intracellularly. Ratio display and statistics are performed live using floating point arithmetic on the raw image data to minimize roundoff error and utilize memory efficiently. Background region can be adjusted after acquisition if spurious signal enters the originally selected background region.
SlideBook allows ratio channels to be collected alongside other fluorescence imaging channels allowing elaborate 3D and 4D experiments incorporating ratio information. It is possible to collect 4D data in GFP and select one plane of each stack to collect Fura-2 data. Ratio imaging can also be part of elaborate experiments which combine ratio acquisition with non-ratiometric labels, 3D and 4D imaging and synchronization with electrophysiology and automated perfusion.
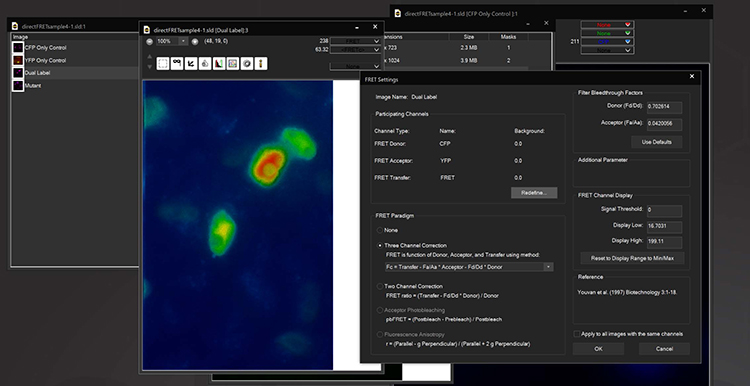
FLIM
The Fluorescence Lifetime Imaging module enables measurement of fluorescence lifetimes via frequency modulation of illumination and detection signals. FLIM is a powerful tool for molecular research in living cells and can be used to measure protein proximity (FRET), polymerization, relative concentration of different molecules, separation of different labels with spectral overlap, ion concentration, and remove autofluorescence. Lifetimes are measured at the sub-nanosecond level by using unique electronic timing circuitry for synchronized phase-shifted illumination. Near single molecule detection is possible. The 3i FLIM system can be used in combination with other multidimensional imaging techniques and images orders of magnitude faster than time domain systems.
TTL Synchronization
The Synchronization (TTL) module provides sophisticated electronic control of carefully timed devices via an electronic controller unit. The TTL controller can both create and receive triggering signals and orchestrate their interaction through an experiment. Timing that is not achievable through standard data connections to a PC is made possible by custom circuitry operating in cooperation with the computer and operating system.
The Synchronization module provides software control of analog/digital I/O boards, utilizing them to direct a variety of signals via TTL and other methods. Key to this functionality is pulse generation and recording. In addition to control of lasers, detectors, shutters and fine motion devices, the module can be used to synchronize external stimulation and data collection instruments such as electrodes, patch clamp recording devices and perfusion systems.

Rapid 4D Streaming
Rapid 4D allows extremely high-speed 3D image capture over time by combining free-run camera streaming with a piezo stage cycling continuously through a z focus range. The output of each camera image is recorded simultaneously with the Z location of the piezo stage. The Rapid 4D module enables both devices to run simultaneously at maximum speed by synchronizing their movements in hardware instead of relying on slower software synchronization. The result is full 3-dimensional z-stacks of data captured at 30, 60 or even 100 stacks per second continuously. This is an order of magnitude or two higher than typical z-stack capture speeds over time. The only limitation to Rapid 4D is camera readout speed and piezo Z performance.

Scanning
The Scanning Module allows SlideBook to control dual galvanometer scanning systems using National Instruments data acquisition hardware. Scanners for such applications as photobleaching, point scanning confocal and multiphoton imaging can be controlled to scan points, lines, and user-defined regions of interest via resonant scanning or standard x,y scanning.
LightSheet
The LightSheet module facilitates control of Lattice LightSheet, Marianas LightSheet, and Cleared Tissue LightSheet alongside light sheet-specific analysis modules such as Localized Cross Correlation Reconstruction, deskewing, channel registration, and deconvolution.

Photomanipulation
The Photomanipulation module allows control of both laser and widefield light sources used for applications such as FRAP, photoablation, and photoactivation. Users can quickly switch from widefield or confocal imaging to photomanipulation to create a dynamic photon event in a region then return to imaging within milliseconds. The Photomanipulation module allows the user to define a region or regions of interest as a single diffraction limited spot or as user-defined shapes. This module is typically paired with Vector for diffraction-limited scanning or with Phasor for simultaneous stimulation of multiple points via digital holography.

Computer Generated Holography
This module controls a phase-only spatial light modulator to produce holographic laser illumination patterns. When used in conjunction with Phasor or Phasor 2-Photon, this module produces simultaneous 3D multipoint and arbitrary 2D pattern photostimulation and photoactivation. Sequences of patterns can be defined and synchronized by time point, real time, or an external digital trigger.
Supported Hardware
| Device Vendor | Device Name | ||
|---|---|---|---|
| Leica | CTR | ||
| Leica | DM4000 | ||
| Leica | DM5000 | ||
| Leica | DM6000 | ||
| Leica | DMR | ||
| Leica | M205F | ||
| Nikon | TE2000 | ||
| Nikon | Ti | ||
| Nikon | Ti2 | ||
| Olympus | BX61 | ||
| Olympus | BX63 | ||
| Olympus | IX81 | ||
| Olympus | IX83 | ||
| Zeiss | Axio Examiner | ||
| Zeiss | Axio Imager | ||
| Zeiss | Axio Observer | ||
| Zeiss | Axio Zoom.V16 | ||
| Zeiss | Axioplan | ||
| Zeiss | Axiovert 200M |
| Device Vendor | Device Name | ||
|---|---|---|---|
| Andor | iXon series | ||
| Andor | Luca | ||
| Andor | Neo | ||
| Andor | Zyla | ||
| Hamamatsu | ImagEM series | ||
| Hamamatsu | ORCA-Flash series | ||
| Hamamatsu | ORCA-Fusion series | ||
| Hamamatsu | ORCA-Quest | ||
| Olympus | DP72 | ||
| PCO | Sensicam | ||
| Photometrics | Cascade series | ||
| Photometrics | CoolSNAP series | ||
| Photometrics | Evolve series | ||
| Photometrics | Kinetix | ||
| Photometrics | Prime series | ||
| Photometrics | QuantEM series | ||
| Zeiss | AxioCam series |
| Device Vendor | Device Name | ||
|---|---|---|---|
| 89 North | Heliophor | ||
| All Vendors | Analog monochromator | ||
| Caim | OptoScan | ||
| CoolLED | pE series | ||
| Excelitas | X-Cite series | ||
| Lumencor | SOLA | ||
| Lumencor | SpectraX | ||
| Prior | Lumen Pro | ||
| Sutter | Lambda series | ||
| TILL | PolyChrome Monochromator | ||
| Zeiss | Colibri |
| Device Vendor | Device Name | ||
|---|---|---|---|
| 3i | LaserStack | ||
| All Vendors | AOTF | ||
| Nikon | Nikon Ti Laser Launch | ||
| Spectral Applied / Andor LMM | Laser Launch | ||
| Uniblitz | VMM-3 |
| Device Vendor | Device Name | ||
|---|---|---|---|
| 3i | Ablate! | ||
| 3i | Nouveau Phasor | ||
| 3i | Phasor | ||
| 3i | Vector | ||
| 3i | Vector2 | ||
| Andor | Micro*Point USB | ||
| Andor | Mosaic |
| Device Vendor | Device Name | ||
|---|---|---|---|
| Any | Any XY stage that accepts analog voltage input | ||
| ASI | MS-2000 | ||
| Ludl | MAC2000-MAC6000 | ||
| Mad City Labs | MicroDrive | ||
| Marzhauser | Corvus | ||
| Marzhauser | LStep | ||
| Marzhauser | Tango | ||
| Olympus | BX3-SSU, IX3-SSU | ||
| Prior | XY Stage | ||
| Scientifica | XY Stage | ||
| Sutter | MPC-200 |
| Device Vendor | Device Name | ||
|---|---|---|---|
| Any | Any Z Stage that accepts analog voltage input | ||
| ASI | PZ and PZU series | ||
| Ludl | MAC2000-MAC6000 | ||
| Physik Instrumente (PI) | Piezo Z Stage | ||
| PiezoJena | Piezo Z Stage | ||
| Prior | Z Stage | ||
| Scientifica | Z Stage | ||
| Sutter | MPC-200 |
| Device Vendor | Device Name | ||
|---|---|---|---|
| ASI | FW series | ||
| Cairn | OptoSpin | ||
| Ludl | MAC2000-MAC6000 | ||
| Prior | HF series | ||
| Sutter | 10-3 series |
| Device Vendor | Device Name | ||
|---|---|---|---|
| ASI | SC-2000 | ||
| Excelitas | X-Cite series | ||
| Prior | Shutter | ||
| Sutter | SC | ||
| Uniblitz | VCM | ||
| Uniblitz | VMM | ||
| Uniblitz | VMM-3 | ||
| Uniblitz | VS |
Partners
Aivia is an innovative and complete 2-to-5D image visualization, analysis and interpretation platform. Using state-of-the-art algorithm and software architecture, Aivia delivers top performance on critical tasks such as display of large images and analysis of complex biological phenomena. Aivia is powered by a range of machine learning technology for both image segmentation, object classification, and novelty detection.
The scripting function in SlideBook can directly control and port data to/from MATLAB. A SlideBook script can export channel and mask data directly to MATLAB without the user needing to access the program. The script can set variables and run a program in MATLAB to analyze the SlideBook data in a MATLAB matrix. Once analyzed the matrix data can be ported back to SlideBook as channels and masks for viewing and supplemental analysis.
Microvolution® software delivers nearly instantaneous deconvolution by combining intelligent software programming with the power of a GPU. SlideBook has seamlessly integrated Microvolution, which offers GPU-accelerated Richardson-Lucy deconvolution of widefield, spinning disk confocal, multiphoton, and light sheet data (including 3i’s Lattice LightSheet) using either a measured point spread function (PSF) or blind estimation. The result is nearly instantaneous deconvolution of data within the application without the need to export to and re-import from a separate application. Microvolution complements SlideBook’s 20+ years of active development of hardware control, capture, post-processing, and analysis tools.
SlideBook fully supports OME-TIFF export for any dataset at the click of a button. In addition, the SLD file format is compatible with any software package using OME Bio-Formats.
Application Data
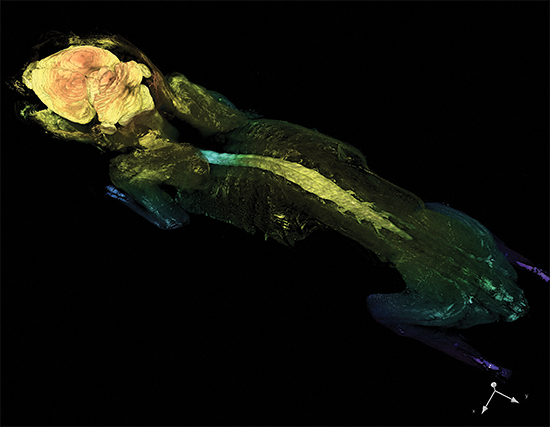
Cleared Tissue LightSheet
Thy1-YFP mouse measuring 4.5cm in length cleared with PEGASOS and imaged with Cleared Tissue LightSheet XL. Sample courtesy of Dr. H. Zhao, Beijing Institute for Brain Research.
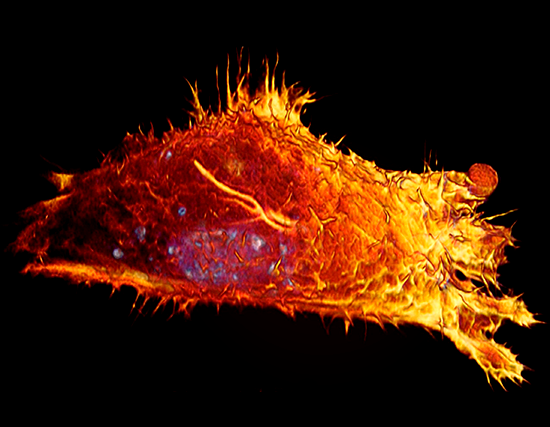
Lattice LightSheet
HeLa cells transfected with GFP-Lifeact (488nm) to visualize actin dynamics. Courtesy of James Springfield, Institute for Molecular Bioscience (IMB) at the University of Queensland.
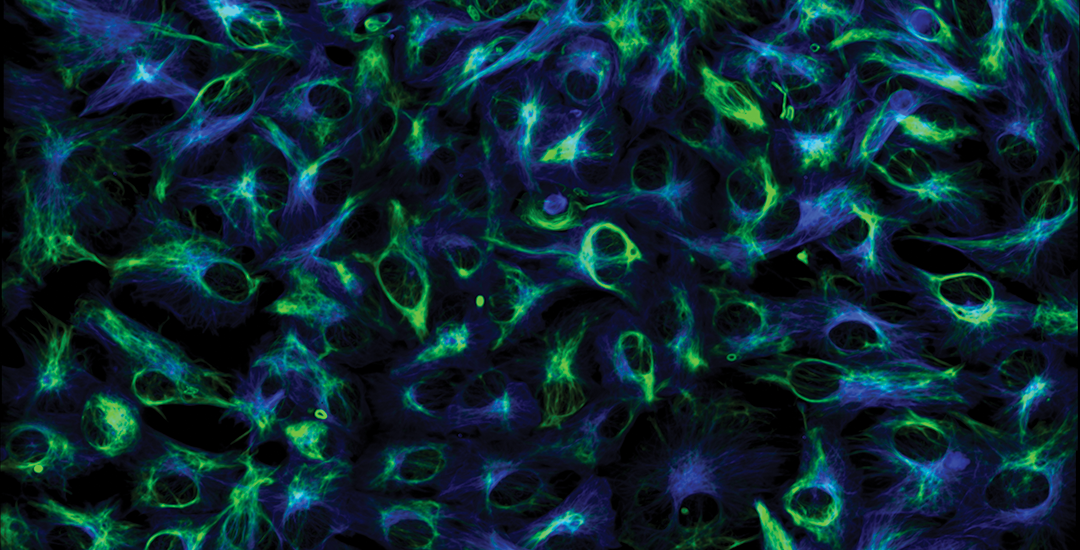
Marianas
Retinal pigment epithelium cells expressing GFP-vimentin (green) and RFP-tubulin (blue). Courtesy of Dr. Doncic Lab, University of Texas Southwestern Medical Center.
VIVO Multiphoton
T4T5 neurons in the Drosophila visual system responding to various directions of visual motion. Courtesy of Dr. Ben Hardcastle, Frye Lab, University of California Los Angeles.
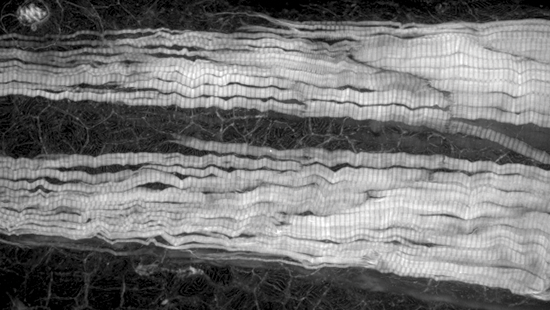
Marianas
Zebrafish tail labeled for actin.
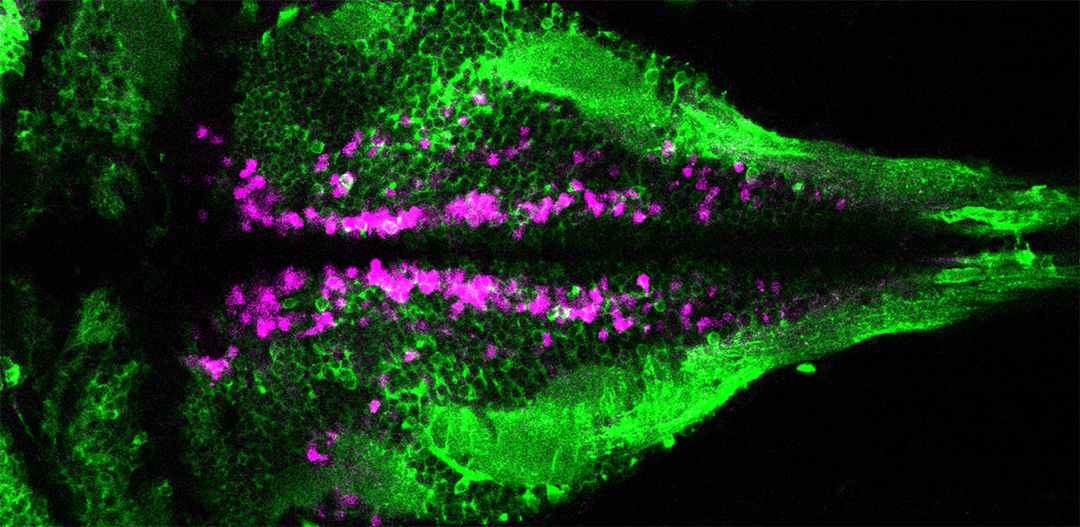
VIVO Multiphoton
A z-stack projection of ciliated cerebrospinal fluid-contacting neurons (magenta) contacting the central canal in the spinal cord of a larval zebrafish. Cilia are labeled in green. Courtesy of Jenna Sternberg, Wyart lab, Institut du Cerveau et de la Moelle Épinière, Paris.

SlideBook Reader is a free download that allows viewing of .sld and .sldy files in the familiar user interface of SlideBook. In addition to viewing data in XY, XZ and YZ projections, Reader includes a handful of useful tools:
- Full renormalization and lookup table options
- Export 16-bit tiff images
- View and graph regions of interest
- Export graphs and graph data
- Perform length and angle measurements
- Display and edit annotations
SlideBook Reader is compatible with Windows 10 and later. MacOS is not available at this time.