Automated, Reproducible and Interoperable Imaging and Analysis
The Synergy module adds a powerful integration layer to SlideBook with a socket-based server mode and robust list of commands for data manipulation, hardware control and more. Synergy enables automated imaging pipelines, precision-controlled hardware operation and seamless interoperability with the open-source tools and computational frameworks used across modern research labs. Designed for imaging facility staff, computer scientists and researchers alike, Synergy brings automation, standardization and open integration to SlideBook’s high-speed high-precision image acquisition and analysis.
Interoperability with Open Ecosystems
Synergy is designed to interface naturally with the tools research labs already depend on, including ImageJ, napari and Neuroglancer. This interoperability ensures data can flow directly into existing analysis pipelines without time- and storage-intensive imports and exports while preserving SlideBook’s robust metadata management.

Create Imaging and Analysis Pipelines
Synergy exposes SlideBook’s data management and hardware control capabilities through a server mode. Using your preferred programming language (including Python, MATLAB, Java, C and more), create autonomous imaging pipelines, incorporate 3rd-party devices and perform AI-based analysis.
Ready for AI and HPC
Synergy enables autonomous control with intelligent decision making as well as integration with AI models and methods. High-performance computing (HPC) servers and workstations can be used to offload resource-intensive processing thanks to Synergy’s socket-based architecture. By exposing core SlideBook capabilities with clear functions and parameters, Synergy is a machine-readable UI designed for next-generation AI interfaces.
Reproducibility and Standardization
Every imaging parameter and hardware action can be scripted, shared, and versioned. This ensures consistent execution across instruments and operators and strengthens the reproducibility and transparency of scientific workflows.
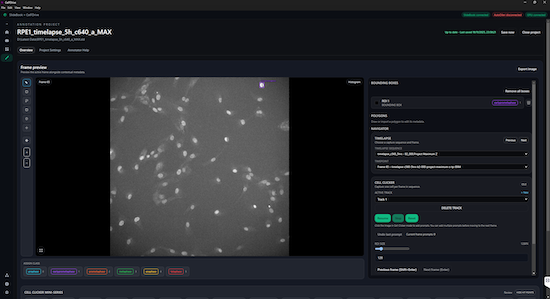
Apps Powered by Synergy
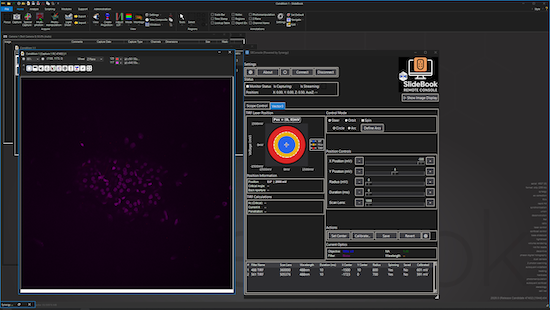
SBConsole
Manage capture scripts, execute multi-script queues, monitor live images, and control hardware components from a single intuitive interface. Build and run complex experiments to automate repetitive tasks. SB Console is ideal for labs looking to implement imaging pipelines, high-throughput imaging, and timelapse or multi-position acquisitions.
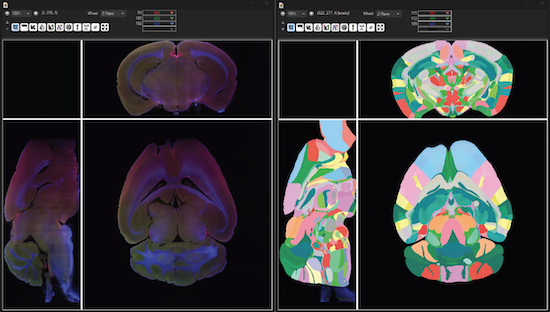
SBAtlas Manager
Import atlases with manual or automated alignment. Atlases are stored as masks fully compatible with SlideBook’s 3D mask quantitation, object measurement and analysis.
Start Building
Use your preferred programming language (including Python, MATLAB, Java, C and more), to harness SlideBook’s data management and hardware control capabilities and start creating autonomous imaging pipelines, incorporate 3rd-party devices and perform AI-based analysis.
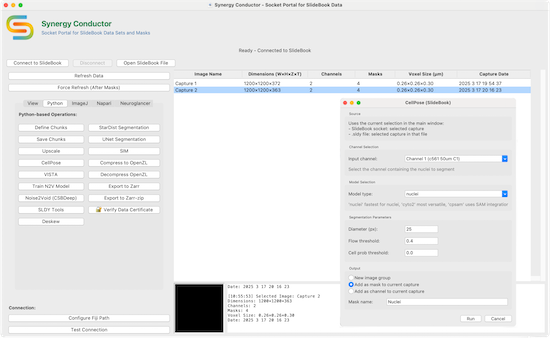
SBConductor
A modular application with an open foundation built on a Python scientific computing stack that seamlessly bridges the gap between SlideBook’s cutting-edge microscope control and acquisition and the latest AI-powered analysis tools and applications. SB Conductor provides a unified interface for advanced data management, processing and verification for local SLD/SLDY files as well as remote access to SlideBook. SB Conductor is cross-platform compatible (Windows, MacOS and Linux) with GPU acceleration support (CUDA, Metal and MPS).
CelFDrive
Solve the “needle in a haystack” problem by programmatically identifying targeted cellular events with low-magnification low-phototoxicity imaging and then autonomously switching to high-speed high-resolution 3D imaging at the correct time, location and duration required. The resulting smart, closed-loop acquisition system boosts imaging throughput, reduces redundant data capture and helps researchers capture more actionable, biologically-valuable data.